PHYTAID: PHYTolith Automatic IDentification
PHYTAID is a tool to automatically identify grass silica short cell phytoliths from 2D microscope images. Fill out the form below and upload an image to get started; the results will be emailed to you within a few minutes. PHYTAID is currently in early testing stage; do not rely on the results yet for publication. We are interested in feedback, please send comments or questions to PHYTAID@uw.edu.Identify your grass silica short cell phytolith with our tool!
About the tool
What is PHYTAID
PHYTAID stands for PHYTolith Automatic IDentification system. PHYTAID uses artificial intelligence, also known as machine learning or a neural network, to classify images of unknown grass phytoliths into one or more taxonomic groups based on a training set of phytolith images from modern grass specimens of known taxonomic identity. The algorithm identifies features from images in the training set that allows it to best classify images into each group. This type of machine learning approach is often referred to as a Black Box approach because it is impossible to set and difficult to extract the actual image features that are being used by the program.
The goal of PHYTAID is to complement the phytolith identification process, which is currently accomplished by human experts, with an unbiased and automated identification method. PHYTAID is currently in its second stage of alpha testing so users should not yet rely upon the results. The current version of PHYTAID is 0.2 and was released on May 1, 2019.
PHYTAID uses a convolutional neural networks (CNN) to identify features through local weighted summations, within small segments of each image. The weights are referred to as filters and each transformation from layer to subsequent layer will usually involve having multiple filters with each filter applying weighted summation onto equally spaced patches from the previous layer. The weighted sums are then stacked together and forms the latter layer. CNN’s with several layers enable early layers to encode local features and latter layers to combine local features to form global features.
Training on 80% of the preprocessed 5,081 images, resulted in correct classification rates of xx%, xx% and xx% at the subfamily, tribe, and sample/species levels respectively.
What types of images can the system accept?
PHYTAID accepts images in .jpg or .tif format or a zip file of multiple images. Images should be taken on a standard bright field microscope using a 100x objective. The phytolith should be in grey scale, unbroken, centered, and without other phytoliths or large amounts of debris in the background. The banner at the top of the home page provides some example images. You can also visit our image library to view the current set of training images used by our PHYTAIDas well as additional grass phytolith images and 3D meshes
PHYTAID Sampling and sample preparation
The grass family consists of approximately 12,000 extant species distributed worldwide. The family is at least 110 million years old and likely originated on the Gondwana supercontinent before spreading throughout the world. Some of the evidence for the age and geographic distribution of grasses comes from the short cell phytoliths produced in the epidermis. While many plant families produce phytoliths grass short cell phytoliths (GSSCP’s) are, as their name implies, only produced by grasses. Fossil GSSCP’s have been found on every continent and the oldest GSSCP’s are at least 66 million years old.
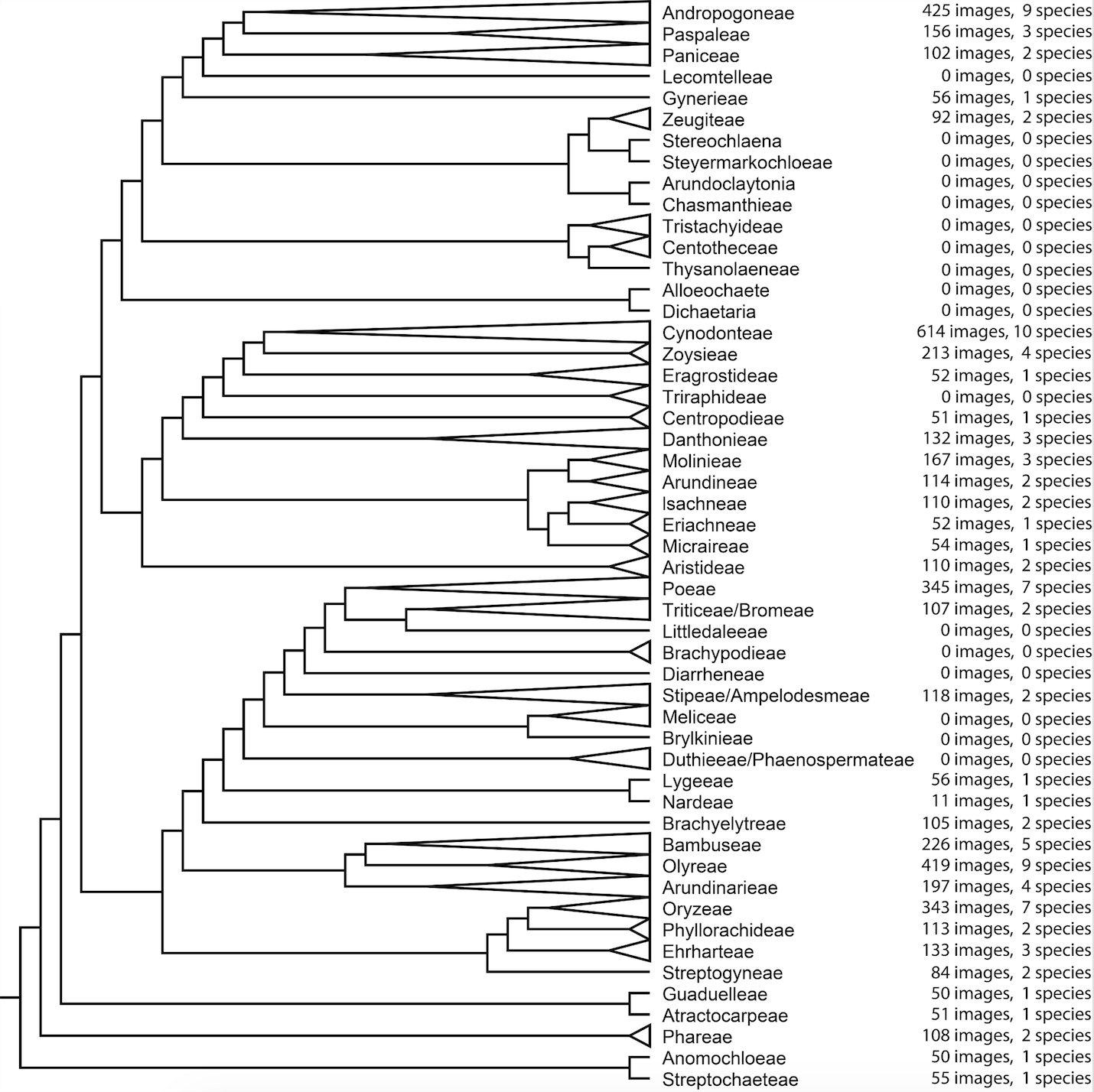
Grass species are classified in subfamilies, tribes, subtribes and genera. There are 12 subfamilies, 52 tribes, 90 subtribes and 768 genera. As far as we know, every grass species produces GSSCP’s. And our observations and the observations of many other phytolith workers suggests that closely related species typically share similar phytolith morphotypes. It would be difficult, if not impossible to represent all the phytolith diversity produced by these species so for the first iteration of PHYTAID we have sampled 100 species representing 65% of tribes. Samples were taken from the Phytolith Modern Reference (PMR) library at the Burke Museum supplemented by material from Iowa State University (courtesy of Lynn Clark and Phil Klahs).
In order to generate samples, leaves are washed and boiled in Schulze’s solution, a saturated aqueous solution of potassium chlorate KClO3 in concentrated nitric acid followed by treatment in hydrochloric acid. This treatment removes organic material and carbonates leaving only silicified material (phytoliths). The resulting residue is placed on a slide and individual phytoliths are imaged. Although the sample may contain other types of phytoliths, only GSSCP phytoliths are imaged.
Future versions of PHYTAID will include higher sample numbers from more species. We aim to have each tribe and subtribe represented by multiple species from each.
How to interpret the results from PHYTAID: important note and disclaimer
There are several challenges that we need to overcome before PHYTAID can move to beta testing and then to a full release. First, PHYTAID is currently not robust to large deviations in imaging conditions, differences in scale or the presence of other phytoliths or surrounding debris captured in the image. The training set of images were taken using a Nikon Optiphot microscope with a PlanApo 100x oil immersion objective and imaged using a 5-megapixel NikonDS-Fi1 camera. Images were converted to greyscale. Phytoliths imaged under different conditions will not yield ideal results under the current implementation of PHYTAID. The image library that was used to train the algorithm is also not comprehensive with regards to the diversity of extant grasses and grass phytoliths. The current training set is based on 5,081 images from all 12 subfamilies, 34 of 52 tribes and 93 of 768 genera. Future releases of the tool will feature an expanded training set and greater robustness to variation in imaging conditions.
The machine learning algorithm will classify any image that it is given. For example, if presented with an image of a cat, the algorithm will give a result classifying the image into one or more grass taxonomic categories. In order to prevent this from occurring we have implemented an uncertainty-aware loss function which rejects inputs when prediction power is poor. A user-adjustable cost parameter allows for tuning the function. At lower values the cost parameter will allow the algorithm to return a set of taxonomic classes and probabilities for an image even if the predictive uncertainty is high. At higher values, any image that is classified with low predictive uncertainty will be rejected and the system will not return a set of classifications. In addition, every classified image will include classification probabilities for each class at each taxonomic level (Subfamily, Tribe, Genus). Classifications with high probabilities in one or two classes at a given taxonomic level should be considered stronger evidence than classification with low probabilities across many classes. Similarly, since classification probabilities at a given taxonomic level are independent of classifications at other levels, when these agree (i.e. the most probable tribe is found within the most probable subfamily) this can also be considered stronger evidence for the classifications.
Shape homoplasy occurs in grass short cell phytoliths This is referred to as ‘redundancy’ in the phytolith literature. This is particularly true from a functional point of view when considering only the two-dimensional shape of the phytolith. Shape homoplasy/redundancy is another potential limitation of PHYTAID which could lead to misclassification. We have imaged the training set of phytoliths as they appear on the slide. Depending on their shape and how they have settled on the slide, the phytoliths for a species may have been imaged at a consistent or at random perspective/rotations.
Once PHYTAID moves from early testing to deployment we will encourage users to, whenever possible, use PHYTAID to identify multiple phytoliths from an assemblage. While a single phytolith successfully classified to a group may be compelling, several phytoliths from an assemblage classified to the same group is stronger evidence for the classification. In addition, most grass species have at least two, often very different, short cell phytolith morphotypes. These typically include a costal (over the veins) morphotype and an intercostal (between the veins) morphotype. In some cases, a species will have more than one costal or intercostal form. This fact can be useful for confirming identifications. While two distantly related species may share one morphotype it is unlikely that they will share two or more morphotypes.
PHYTAID is currently still in development and has been made available for testing purposes only. Use PHYTAID at your own risk. The PHYTAID team, the University of Washington and the Burke Museum shall not be held responsible or liable, whether directly or indirectly, for any damages or loss caused or sustained by the user in connection with any use or reliance on information or classification results obtained by PHYTAID or this website.
What is a phytolith?
Phytoliths are microscopic silica bodies that precipitate in and around cells in many plants. When plant tissues decompose, their phytoliths are deposited in the soil, sediment, or archaeological context. Study of phytoliths is important in archaeology, where it has led to discoveries about the diet and plant use by humans (e.g., domestication of crop plants), and in paleobotany, where it has allowed the reconstruction of plant diversification and vegetation change many thousands or millions of years ago.
Most major groups of plants contain species that produce phytoliths, and grasses (family Poaceae) deposit more silica in their tissues than most other groups. A particular kind of phytolith that is unique to grasses are the so-called grass silica short cell phytoliths (GSSCP). They form in specialized cells on the surface of the plant (the so-called epidermis) called silica short cells and take on a wide variety of forms. The shapes of the GSSCP are often unique to particular grass genera or groups, and archaeologists or paleontologists routinely rely on the shapes of GSSCP in their samples to infer what types of grasses were being used or that lived in an area.
However, although GSSCPs can differ among grass groups, there can also be some overlap among distantly related grass taxa, making it hard for the human eye to distinguish and correctly determine which taxon a particular GSSCP shape (“morphotype”) comes from. This is where the PhytAID tool comes in. It uses Machine Learning & Computer Vision to establish the most likely classification for a morphotype.
Further reading:
Piperno, D. R. 2006. Phytoliths: A comprehensive guide for archaeologists and paleoecologists AltaMira Press, New York.
Piperno, D. R. 2014. Phytolyth Analysis: An Archaeological and Geological Perspective. Academic Press, San Diego, CA.
Strömberg, C. A. E., R. E. Dunn, C. Crifò, and E. B. Harris. 2018. Phytoliths in paleoecology: Analytical considerations, current use, and future directions. Pp. 233-285. In D. A. Croft, S. Simpson, and D. F. Su, eds. Methods in Paleoecology. Reconstructing Cenozoic terrestrial environments and ecological communities. Springer Publishers.
How to cite PHYTAID
For PHYTAID
PHYTAID (2019). “Phytolith Automatic Identification System” v.0.2. Facilitated by the University of Washington and The Burke Museum of Natural History and Culture. Published on the Internet; http://www.phytaid.burkemuseum.org/ Retrieved dd month yyyy."
and
Perry, J, Meng X, Zhang J, Gallaher T, Jamieson K, and Strömberg C.A.E. Automated Classification of Grass Phytoliths Using a Machine Learning and Computer Vision Approach (In prep)
and for the uncertainty-aware loss function
Anonymous and Anonymous 2019, Uncertainty-aware black-box predictors with coverage guarantees [Under review by the International Conference on Machine Learning (ICML)]
Privacy policy, your data and personally identifiable information
All information submitted to PHYTAID will remain confidential.
We record the names, email addresses and affiliations of all user to track usage but personally identifiable information will not be shared with anyone outside the PHYTAID development team.
Images submitted by users remain the property of the user. By submitting the image to PHYTAID the user authorizes the PHYTAID team to retain a copy of the image and the resulting classification for purposes of evaluating and improving the tool.
Images and classifications will not be shared with anyone outside the PHYTAID development team without obtaining the consent of the user. Questions or comments regarding this privacy policy should be addressed tophytaid@uw.edu.
The PHYTAID Development Team
Caroline Strömberg, Timothy Gallaher, Kevin Jamieson, Jessica Perry, Xiangyun Meng, Jifan Zhang.
With thanks to Peter Li, Patrick Spieker, Sophia Druet, and Claire Grant.
Web UX/UI, development and design by Lorraine Sawicki, Calyxia LLC
We thank the University of Washington Royalty Research Fund for awarding a grant for the initial development of PHYTAID